The Digital Salmon – Key to both More Science and More Profit
Introduction
Norway has been a prime contributor to the development of today's knowledge base for aquaculture-related salmon biology, and this knowledge base has been
essential to the industrial success of Norwegian salmon farming. Genomic information is a key premise for advanced biological research and development, and
Norway is now playing a major role in the international collaboration to sequence the Atlantic salmon genome. However, access to a high-quality genome
sequence is only a first step towards establishing a sorely needed quantitative and widely applicable knowledge base for one of Norway's most important
export articles. The construction of such an intellectual edifice exposes us to the most daunting challenge facing modern biology, namely how to build a
causal bridge between the genome, environmental influences, and the phenome (the totality of an individual's measureable traits). In line with current
international trends within biomedicine and production biology, we argue that the only feasible way to establish such a knowledge base is to aim for the
construction of a Digital Salmon – a comprehensive, computer-encoded representation of our growing knowledge of salmon biology.
Patterned on similar endeavours in human biomedicine, a Digital Salmon would encompass an ensemble of mathematical descriptions of salmon physiology,
combining mathematics, high-dimensional data analysis, computer science and highly advanced measurement technology with genomics and experimental biology
into a concerted whole. The development of such a quantitative framework by merging life sciences, mathematical sciences and engineering would be highly
instrumental in adapting salmon to new and sustainable feeds without sacrificing product quality, in understanding complex diseases and developing new
vaccines, and in preserving wild salmon populations. In addition, it would contribute strongly to the visibility of Norwegian biological research at the
international research frontier while providing the foundation for a new biotechnological industry.
It takes all the running you can do to keep in the same place: The future of salmon research and development in Norway
The Red Queen is a fictional character in Through the Looking-Glass (Carroll, 1871), the sequel to Alice's Adventures in Wonderland. Having led Alice to the top of a hill, the Red Queen begins to run, faster and faster. Alice
runs after, but is baffled to find that neither one seems to be moving. Alice remarks on this, to which the Red Queen responds: "Now, here, you
see, it takes all the running you can do to keep in the same place". This is an apt metaphor for what it takes to sustain a long-lasting and
profitable Norwegian salmon industry.
To maintain production volumes, not to mention move forward, this industry has to continuously deal with and adapt to a range of biological and
non-biological challenges that make up a demanding Darwinian theatre where the possibility of severe decimation, or even extinction, always lurks behind
the scene. The salmon industry stands out among Norwegian knowledge-driven export industries by having only a rudimentary understanding of how its product
is designed and how to adjust this design to the demands of production or market conditions. This is due to the fact that the product is a highly complex
organism, kept in an uncontrolled environment (the sea), where major biogenic production factors (feedstuffs) must be acquired from highly competitive
markets. Furthermore, even though the industry has successfully dealt with several of the challenges it has been exposed to during its 40 years' history,
the predicament laid out by Red Queen is unavoidable. Therefore, there is a need to actively acquire and utilize a deep knowledge of salmon biology; a need
that will become more and more prominent as the industry expands.
Recently, the Norwegian academies of science commissioned a report titled Value created from productive oceans in 2050. Looking forty years into
the future, the authors foresee that "In 2050 biotechnological and genetic methods have enabled the development of a sterile salmon with excellent growth
properties and robust immune system" (Olafsen et al., 2012 p.44). Without debating the merits of this goal, we acknowledge that a sterile salmon
is within reach long before 2050. However, the claim that biotechnological and genetic methods have enabled the development of a salmon with excellent
growth properties and robust immune system, is arguably not paying enough attention to what is conveyed by the above Red Queen metaphor. In this chapter,
we address what it takes to develop and maintain a salmon with excellent growth properties and a robust immune system while the estimated annual production
increases from the current 1 million tons/year to a prospected 5 million tons/year in 2050 (Olafsen et al., 2012). Our major claim is that this
goal is not likely to be achieved in a sustainable way unless we take Norwegian research and development on salmon biology to a completely new level along
several theoretical, experimental, technological and organizational axes.
The need for a systems understanding of salmon biology
Norway is at the fore of the international collaboration to sequence the genome of the Atlantic salmon, thanks to co-operation between Norwegian research
groups, the industry and the Research Council. Genomic information is a key premise for advanced biological research and development, and the salmon genome
is expected to yield unique knowledge of relevance to great current challenges in aquaculture as well as the management of wild salmon. In fact, DNA
information is used in at least four types of application in biology (Gjuvsland et al., 2013):
-
As a pure marker of phylogenetic origin and history, without reference to any particular phenotype (i.e. observable trait of
interest). Examples include quantifying genetic divergence between salmon stocks from different rivers, identifying escaped farmed salmon, studying
selection history and effective population size, and describing genome structure.
-
To detect statistical association between variation in DNA and in phenotypes. Salmon phenotypes currently being targeted are for
example growth rate, colour, fatty acid composition, the ability to utilize different feeds, and resistance to disease agents.
-
To document the existence of a causal connection between a particular DNA variation (natural or imposed) and variation in a specific
phenotype. For example, many single-nucleotide polymorphisms (SNPs, i.e. point mutations) may all be correlated with a given
phenotype, but only a few may have a causal effect.
-
To account in a causally cohesive way for the genome-phenome relationship in terms of biophysical mechanism at the cell, tissue and
organ system levels under various environmental conditions and life stages. We will specify what this means by defining some key concepts. The term system denotes a set of interacting and interdependent components that form a unified whole. In particular, living beings are dynamical systems: their state changes over time due to the physiological processes of its component cells, tissues and organs. The adjective
"cohesive" means "causing to cohere" (Merriam-Webster, 2008), and Rajasingh et al. (2008) defined a causally cohesive genotype-phenotype model as one
that, at some given level of resolution, has the quality of causing components involved in a genotype–phenotype relation to cohere in a logically
consistent and ordered way. This is in contrast to standard population genetic models, where phenotypic values are assigned directly to genotypes
without involving any intermediate processes.
Thus, fish are dynamical systems in that physiological processes change the state of the body, and the rates of the various processes depend on the body
state and external influences. These dependencies may vary for both genetic and environmental reasons. Odd Arild Olsen’s Dr. Ing. thesis at the Norwegian
Technical University more than 20 years ago, titled Structured modelling of fish physiology, represents a truly pioneering contribution in this
direction (Olsen, 1989).
Using statistical associations, Norwegian breeding programmes for salmon are now successfully selecting individuals resistant to IPN (infectious pancreatic
necrosis) even before the full salmon genome is available (Anon, 2010). A few research groups have also already embarked on the much more challenging task
to identify the specific causal genetic variation underlying important traits. Even though these two approaches are likely to be sufficient for tackling
several important issues in the years to come, it should be kept in mind that their usefulness depends on the availability of existing genetic variation in
the salmon population. Due to this they provide only a very narrow window of insight concerning the genome-phenome relationship as such, an insight that is
needed for establishing a really pre-emptive knowledge base providing comprehensive explanatory and predictive capability. Only a causally cohesive
understanding can meet this need, and this is what a Digital Salmon will provide.
The genome-phenome relation is the outcome of very complex dynamics that are also heavily influenced by the environment. It cannot be overemphasised that
there is no direct causal arrow from genotype to phenotype in the sense that DNA is responsible for exerting a direct effect as a sub-system on
the system dynamics. The causality flows from the physiological state of the organism through a change in use of DNA, which in turn influences the
physiological (or systems) state. In those cases where variations in DNA cause changes in the system dynamics, it may lead to detectable physiological
variation (Omholt, 2012). This way of perceiving the function of DNA in a genome-phenome context brings quantitative physiology back as the major arena for
understanding organisms.
The standard definition of physiology is “the study of the functions and activities of living matter (as of organs, tissues, or cells) as such and of the
physical and chemical phenomena involved” (Gove, 1981). Thus it is a discipline that per definition seeks integrative understanding of the body as a system. Such understanding is exactly what we need if we are to change the fact that one of Norway’s largest export industries has only a
rudimentary understanding of its product. And it will not be obtained unless we start making use of the vocabulary designed for describing and analysing
dynamical systems, namely mathematical modelling. Biological systems are just too complex to be understood in systemic terms by the simple conceptual
models still used by most biologists. Despite their usefulness, the same is true for the sophisticated statistical models and bioinformatics tools
currently being employed to extract understanding from the huge –omics data sets that are accumulating at an increasing rate. (The suffix -omics is used in
biology to form nouns meaning a study of the totality of something, e.g. genomics, proteomics; Wiktionary, 2013.)
The efficacy of computational models in physiology arises from their capacity to connect a comprehensive amount of empirical data into a functional whole:
-
By enforcing explicit formulations of various hypotheses.
-
By precisely framing the quantitative consequences (prediction space) of hypotheses, by initiating and canalizing experimental or empirical work
by pointing out key questions and the type of data needed.
-
And last but not least, by providing a very efficient interface for communication between a whole range of disciplines with only partly
overlapping vocabularies, helping to integrate the intellectual capital of these disciplines.
These are the rationales for the large investments now being made worldwide to mathematize biological research.
Two of the most important long-term requirements for the salmon industry are the following. (i) It needs to be able to compose a low-cost diet of
sustainable food items, obtained from a highly dynamic global market, that does not compromise the salmon’s physiology. (ii) It needs to understand the
disease physiology of salmon associated with pathogens and parasites so well that when a new epidemic arises, one will, if biologically possible, be able
to fast and efficiently remedy the situation and give the salmon a long-term competitive edge. It is wishful thinking to assume that this will be achieved
without the massive involvement of advanced mathematics, high-dimensional data analysis, computer science and new advanced measurement technology. Such an
endeavour will be outlined in the following sections.
The Norwegian salmon production industry qualifies as a biotechnological industry under the definition used by the OECD and Norwegian authorities:
"the application of science and technology to living organisms as well as parts, products and models thereof, to alter living or non-living materials for
the production of knowledge, goods and services" (OECD, 2005). A merging of life sciences, mathematical sciences and engineering to develop a
transformative knowledge base for the Norwegian salmon industry concords with major trends in international biotechnological research and development. In
the following we outline international developments on establishing mathematically phrased and biophysically based understanding of human physiology and
illustrate how Norway can piggyback on this development to make its own stamp at the international fore through a vision-based and pre-emptive R&D
biotechnology programme aiming for a Digital Salmon.
Piggybacking on the Virtual Physiological Human
The Virtual Physiological Human (VPH) is a pan-European effort
that aims to "revolutionize twenty-first century bioscience by fundamentally shifting the basis for the diagnosis and treatment of disease" by
"biophysically based computational modelling of the human body and physiology" (Hunter et al., 2013). Solid EU funding under FP7 is a recognition
that specialization in medicine is reaching its limits with regards to drug discovery, health budgets and identifying the treatment that best fits each
patient. The vision of the VPH is therefore that "Medical innovation should … be directed towards optimizing treatments using … a customized computer model
of the patient's condition across multiple organ systems and length scales, and across time and environment" (Hunter et al., 2013). Developing
this Digital Patient will involve both the doctors that deal directly with patients, applied research on populations, development of new drugs, and basic
research on physiological understanding and mathematical modelling.
The VPH builds on the worldwide Physiome Project,
and its framework for modelling the human body using biochemical, biophysical and anatomical information on cells, tissues and organs (Hunter & Borg,
2003, www.physiomeproject.org; see Figure 1). Each organ has its subproject, describing for example gas flow in the lungs, mechanics for the skeleton,
electrophysiology for signal propagation in the heart and nervous system, soft-tissue mechanics for muscle contraction, and biochemistry for metabolism. A
major challenge is that a functioning organ relies on an interplay between fast and slow processes on micro, meso and macro scales. Hence, submodels are
developed to describe processes on different levels of organization from single cells to tissues to the whole organ, and these submodels are coupled and
integrated into multi-scale and multi-physics models. The Digital Patient reflects a current push to represent variation between individuals. Some
challenges are only beginning to be handled, such as the organism-level interplay of organs, how environment modifies the genotype-phenotype relation, and
the robustness and plasticity of bodily development.
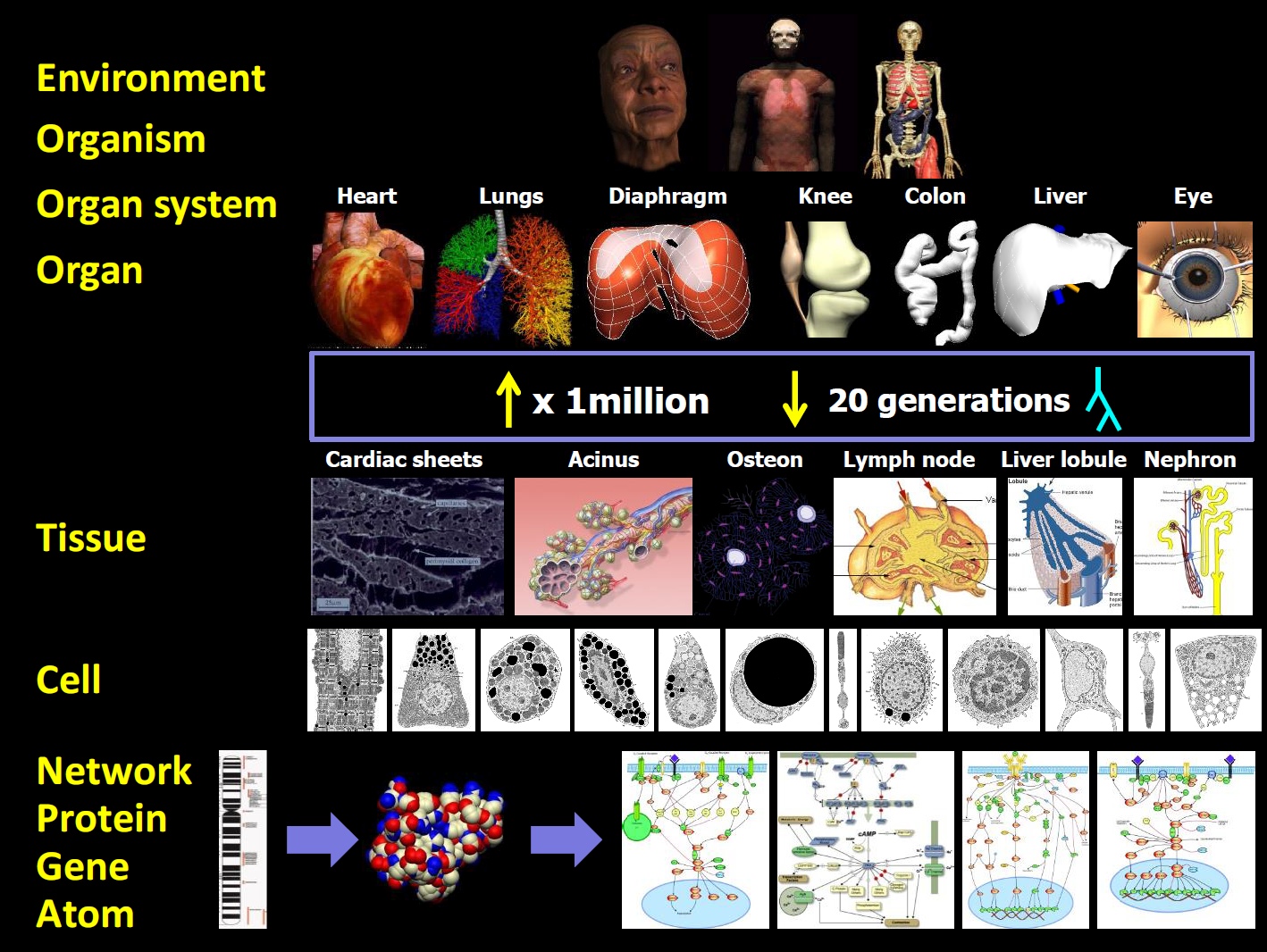
Figure 1. The Physiome challenge: From organs to proteins across enormous scale differences in time and space. Figure courtesy of Peter J. Hunter.
The Physiome Project represents an unprecedented scaling up
of the use of mathematical modelling to consolidate and integrate physiological knowledge, a wide array of observable quantities throughout the phenotypic
hierarchy and physical laws and principles, into comprehensive structure-function representations of cells, tissues and organs. This
advanced engineering approach makes use of some of the most cutting-edge technology and theoretical constructions currently available. Models of this kind
provide an ideal framework for exposing our understanding of how the body works to confrontation with high-resolution biological data.
The Physiome Project has developed a strong infrastructure
to integrate the data and knowledge needed (Figure 2). Model repositories store mathematical models in a format that allows both mathematical analysis,
autogeneration of computer code, and precise labelling of the components of the model. This labelling uses standardized vocabularies for biological
knowledge, making it easy to find models for a particular phenomenon or data to validate a given model. Researchers can ask biologically meaningful questions that get translated to machine-processing of enormous amounts of data, including databases of genome sequences, gene
expression and protein function.
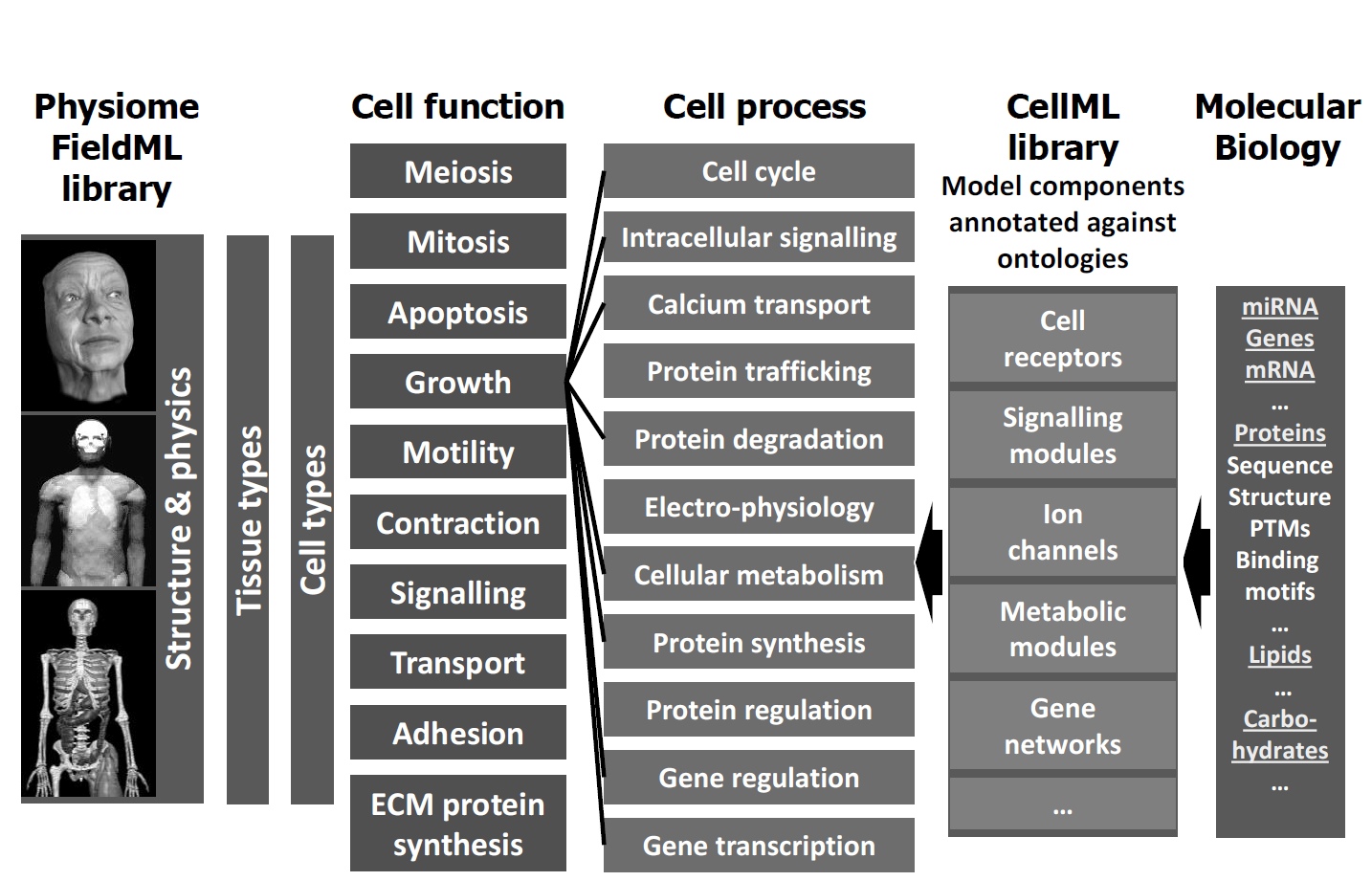
Figure 2. Infrastructure for linking molecular biology to physiome. Figure courtesy of Peter J. Hunter.
Model markup languages
such as CellML and SBML are used to encode mathematical models, similar to how HTML encodes webpages. They have made it much easier to check, share and
reuse mathematical models. This is essential for testing and refinement, especially in integrative work such as virtual physiology. Moreover, a common
language makes a model usable with software for many different purposes: human-friendly formatting, automatic checking that units match up on both sides of
an equation, mathematical analysis, or generation of computer code for numerical simulation. This is a huge improvement from the slow, error-prone process
of coding models from scratch based on a textual publication.
Model repositories
are databases of mathematical models in a standard coding. Models are curated to verify that they reproduce results from the paper in which they
were published, and to ensure standardized labelling of model components, variables and parameters. The major repositories today (biomodels.net and
cellml.org) have a few hundred models each, mostly for differential equation models of cell biology such as metabolic networks or cellular
electrophysiology. This is likely to expand fast in the near future, both in the number of models and the breadth of biological processes and mathematical
frameworks, through projects such as Drug Disease Model Resources (ddmore.eu).
Ontologies.
The vast knowledge embodied in genomic and phenomic databases and model repositories can only be used effectively together if data and models are properly
labelled, allowing data and model resources to speak the same language. This requires formal representations of knowledge, called ontologies,
which are lists that uniquely identify concepts and the relations between them within a specific domain (de Bono et al., 2011). Thus, labels such
as "abnormal enlargement of liver cells" can be given a precise technical meaning, allowing consistent exploration and querying of multiple data and model
resources by bioinformatics means. Even data that is private (patient details, intellectual property, or preliminary work) can be publicly labelled, thus
making potential users aware that such data exist, after which access can be negotiated. Such labelling and cross-referencing is already in place for many
-omics databases, whereas for model repositories it has just started. Ontologies exist for anatomy, medical terms, gene function, protein structure, and so
on. The BioPortal at bioontology.org gives a human-friendly overview (Noy et al., 2009).
Phenomics
denotes the systematic, massive measurement of phenotypes such as gene expression, metabolic activity, anatomic structure from the subcellular to the whole
body, and response to diagnostic stimuli (Lanktree et al., 2010; Houle et al., 2010). This is still rare and mostly limited to model
organisms such as fruit flies or mice (Houle et al., 2010), but will need to become routine to realize the ambitions of physiome research and
human medicine (Gurwitz, 2012; Hunter et al., 2013). Furthermore, phenomics must go hand in hand with modelling, both to prioritize what to
measure and to meet the main challenge of -omics research, namely that of integration and meaning (Joyce & Palsson, 2006). This poses challenges and
opportunities for research funding and industrial innovation alike (Hunter et al., 2013).
Scurrying in the footsteps
of the Virtual Physiological Human comes the Virtual Physiological Rat (Beard et al., 2012), a large systems medicine
project linking genetics to models of the cardiovascular system. Similar efforts for the mouse (Land et al., 2013) and other model organisms are
underway; for instance, the zebrafish has become an important model organism for disease and the immune system (Lieschke & Currie, 2007; Weinstein et al., 2009).
The Digital Salmon
A flexible basis of knowledge is necessary for rapid response to shifting circumstances in aquaculture.
When a new challenge arises, we must be able to easily reanalyse our existing information and understanding of salmon biology. If information is found to
be lacking, we must be able to rapidly acquire new data and incorporate it straight away into a unified whole. To deal with upcoming issues in combating
disease or adapting feed composition while ensuring product quality, we need to understand the interplay between physiological processes (including the
effects of genetic variation), the state of the body, and external factors such as feeding, pathogens and temperature. In this way, we shift from a reactive to a pre-emptive R&D strategy.
A Digital Salmon would encompass a coherent digital and mathematical representation of the salmon body, which can be adjusted to
represent individuals with different genetic make-ups, populations, disease states, feeding regimes, or other scenarios of interest. Pragmatically, it will
mean a suite of computer models for specific purposes, with different simplifying assumptions and levels of detail. Crucially, these models will be linked
to databases of biological knowledge, using human-friendly systems for biologically meaningful queries across many data sources at once. Tailored user
interfaces will serve different user groups in academia, the institute sector and industry and different activities (e.g. developing or refining
mathematical models, using established models for management decisions and experimental design).
The integration of physiological knowledge in a Digital Salmon would address many of the challenges identified in the introduction. It would serve as a
common framework and intellectual meeting place for a wide array of various competences in a similar manner to the Virtual Physiological Human. Most of
these competences in life sciences, mathematical sciences and engineering are currently not involved in research on fish physiology, disease, nutrition,
and product quality. We strongly believe that this extended involvement will make scientific progress faster and more adaptable to the needs of the moment,
bringing together modelling and measurement, theory and experiment. The effect of feed composition on health and meat quality could be followed through the
physiological hierarchy via protein synthesis, metabolic regulation, and growth processes. Shifting feed availability could be met by a coupled
optimization of metabolism and economic considerations, based on rapid iteration between feeding experiments, parameter estimation, and growth simulations.
The fight against new diseases would be guided by a systems understanding of immune responses. For example, candidate vaccines could be tested under the
scrutiny of massive phenotyping, revealing what distinguishes an effective response and suggesting new options.
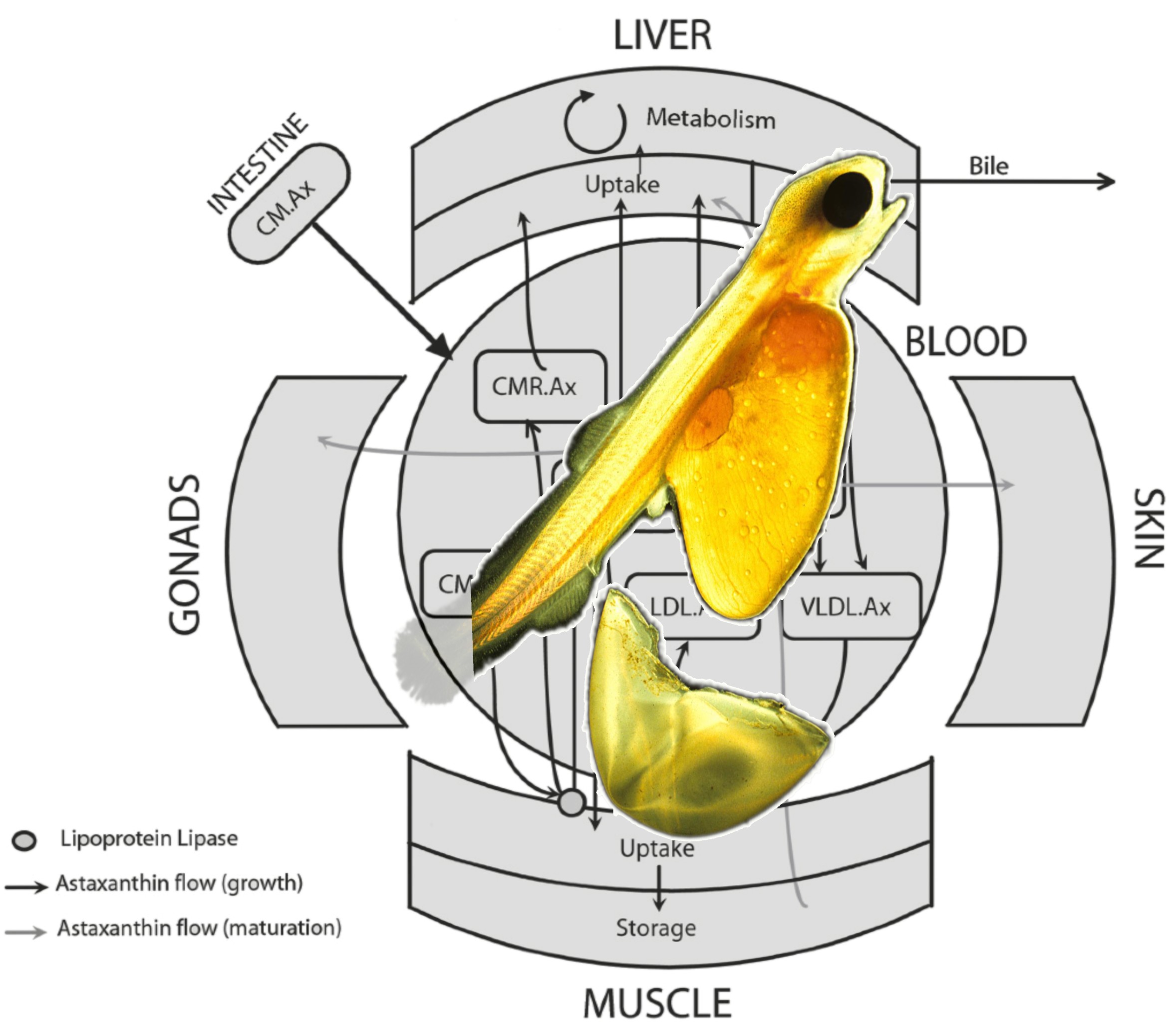
Figure 3. The Digital Salmon is a vision of a coherent digital and mathematical representation of the salmon body, its organs, processes and functions.
Piggybacking on similar efforts in human biomedicine, a Digital Salmon would bring together vast amounts of physiological knowledge in a quantitative,
predictive whole. Such a knowledge structure would make for much more rapid and flexible responses of salmon farming to novel challenges and shifting
demands in disease, feed availability and pricing. The diagram illustrates salmon meat redness (a commercially important phenotype) from a systems point of
view. The red pigment astaxanthin (an expensive component of the feed) is taken up from the intestine, transported in blood and deposited in muscle, skin
and other organs. The rates of these processes depend on the body state (e.g. sexual maturation: grey vs. black flux arrows) and external influences (e.g.
diet composition). Rajasingh et al. built a biophysically based mathematical model to mimic pigment uptake and retention over short and long time periods
(hours, days and years) for various feed experiments, shedding light on individual variation in pigment utilization and meat colour. (See Acknowledgements
for image credits.)
Much of the infrastructure for the Digital Salmon can be lifted directly from the Virtual Physiological Human
and the Digital Patient, in particular the standard formats and ontological labelling systems for data and model resources, and "best practices" for the
development and use of such resources. However, although there is a long tradition of using existing models and data for other species as a starting point
(Niederer et al., 2009), much of the species-specific data and knowledge remains to be developed, and this will require both spadework and
innovation.
Phenotyping programmes for salmon
will have a strong starting point in existing Norwegian expertise, as built up in the industry, the institute sector and the universities. But the current
activity will have to be dramatically expanded and much more concerted. Two important areas of further development are systematic acquisition of -omics
data (gene expression, proteomics, metabolomics, etc.), and the linking of these data and massive amounts of other types of phenotypic data to mathematical
model parameters and outputs. In a very important sense, parameters are phenotypes too (Vik et al., 2011). Because models differ in their emphasis
and level of detail, what is treated as a given parameter in one model could be the output of a more detailed submodel. Moreover, many model parameters
show important individual variation (e.g. genetic variation in fatty acid and protein metabolism and its sensitivity to diet, Morais et al.,
2012). Thus, parameters in physiological models are phenotypes linked by a causal mathematical structure (Gjuvsland et al., 2013); succinct and
relevant summaries of the consequences of underlying mechanisms for higher-level physiology. In most cases we do not yet have technology for measuring such
parameters efficiently. Current ‑omics data cannot be used in a direct way for this purpose, and the linking of such data to models is currently a theme at
the research frontier of bioinformatics and high-dimensional data analysis. Focusing on the needs of physiome modelling will drive the construction of new
mission-critical measurement technology and new bioinformatics of generic value as well as guide experimental work, greatly increasing the value and
generality of the results obtained.
Salmon farming in the future must navigate many conflicting demands:
maintaining a product that can command top prices in the fish market, making fish grow efficiently on whatever sustainable feedstuffs are available while
staying healthy and producing meat with good properties of taste, texture, nutrition and storability; and also dealing effectively with disease challenges.
Just as in the Red Queen story, those who stand still will be left behind. This means that there will be no fixed optimal strategy, but rather a strategy
for quick adjustment of diet within constraints of price and quality, or rapid development of countermeasures in the face of novel diseases. Such
flexibility requires predictive, quantitative models based on integrated knowledge of physiology. In the following we outline four possible applications of
the Digital Salmon.
Fish meal is increasingly replaced by plant-based protein sources for reasons of sustainability and economy (Naylor et al., 2009). However, this
poses great nutritional challenges, especially for a carnivore such as the salmon (Barrows et al., 2008). Plant feeds have less protein, more
starch and fibre, and unfavourable amino acid and mineral profiles (reviewed in Torrissen et al., 2011). Supplementing the feed with vitamins and
amino acids which are lacking in plants often fails to have the intended effect. Protein synthesis may increase, but so does the catabolic breakdown of
protein (Wacyk et al., 2012). It seems the body responds differently to the crystallized amino acids added to feed than to natural feeds.
Furthermore, many plant compounds act as antinutrients for salmon, interfering with the absorption of other nutrients (Krogdahl et al., 2010). A
prime example is soybean meal, which is well tolerated by many fishes but turned out to cause inflammation of the gut in salmon. Even though the feed
industry alleviated this problem by developing soy protein concentrates, we are still facing the fact that current R&D activity on feed development is
to a large degree based on a very costly and time-consuming trial and error strategy. A knowledge base allowing reliable prediction of how salmon
physiology will respond to a combination of feeds is still a long way off.
In 10 years, a typical salmon feed has changed from containing 10 ingredients including one protein source, to more than 30 ingredients including several
protein sources. The feed industry systematically monitors the cost and availability of hundreds of potential feed ingredients and uses this information to
minimize production costs while staying within the bounds demanded by physiology. With so many alternative feed ingredients, it is clear that it is grossly
inefficient to continue identifying the physiological bounds by specific feed experiments only. We sorely need to establish a systems understanding of
metabolism through a Digital Salmon Programme, from which we can predict the physiological consequences of various feed recipes with a minimum of
experimental effort.
By domesticating the salmon to mostly a plant diet the salmon industry will escape the accusations of depleting the oceans, but it will not escape the fact
that it then has become an ordinary terrestrial meat production industry. As the increasing global meat production is considered by the Food and
Agriculture Organization of the United Nations (FAO) to be one of the main drivers behind climate change and loss of biodiversity (FAO, 2006), we should,
for several reasons, seriously start to think about developing more environment-friendly solutions for producing salmon feed by advanced industrial
biotechnology. One possibility is to start using bacterial protein meal grown on natural gas (Øverland et al., 2010). This solution, which is
arguably a sustainable solution when we consider the impact of food production in a wider environmental perspective, is already technologically mature. And
a large-scale industrial implementation standing on the shoulders of many years of Norwegian R&D is now under consideration in Malaysia. Furthermore,
even though we have known for 25 years that we would run out of naturally available fatty acids, this challenge is only now becoming seriously addressed by
the industry and R&D sector. To remedy this extremely serious problem, we either have to produce fatty acids at an industrial scale by novel methods or
design a salmon capable of synthesizing these acids. A change to predominantly novel diets produced by means of industrial biotechnology, would strengthen
the need for a quantitative systems understanding of salmon metabolism.
The purpose and function of the metabolic system is to adjust the rates of the physiological processes of building up and breaking down, in such a way as
to serve the body's needs depending on the state it is in. The model repositories at cellml.org and biomodels.net already contain numerous models of
metabolic signal processing and metabolic pathways. The former describe the rapid process of sensing physiological state (e.g. pH, iron concentration, or
metabolite concentrations) and propagating the signal (e.g. through conformational switches of enzymes) so as to adjust metabolic activity. The latter
describe the longer-term effects of that activity, using the rules of protein physics, mass conservation and chemical stoichiometry. This provides a
framework for understanding how metabolic regulation can go awry if the effect of artificial feed components differs from that of their natural
counterparts (e.g. Wacyk et al., 2012). An organ model for the gut should be a high priority for the Digital Salmon.
The models and phenomics of a Digital Salmon would pave the way for a predictive, well-tested model of metabolic response and growth performance for a
large space of possible diets at the level of the individual fish. Once validated, such a model would become a tool for the industry for rapid optimization
of diet composition based on feed prices and requirements for efficient and healthy fish growth. Further refinement of the model or its parameter estimates
could be a task for basic and applied research, respectively.
Maintaining product quality
Diet is also important to the quality of the final product. For example, rainbow trout got fatter on soy than on fishmeal when fed amino-acid-balanced
diets with either soy or fish meal as protein source (Wacyk et al., 2012). Even so, the proportion of healthy omega-3 fatty acids in salmon
fillets was lower when plant oils replaced fish oils in the feed (Torrissen et al., 2011). Meat redness is another key phenotype, resulting from
the uptake, transport and deposition of carotenoid pigments in different tissues; Figure 3 illustrates a model of how these processes depend on sexual
maturation (Rajasingh et al., 2006). Because carotenoids are chemically similar to fatty acids, this marks a first step towards systems modelling
of fatty acid metabolism in salmon. Understanding the relationship between feed composition and product quality requires additional emphasis on models for
describing a wide set of biophysical muscle tissue characteristics of relevance to meat quality. The Digital Salmon will then delineate the regions in
"diet space" that ensure satisfactory product quality. This requirement then forms a constraint for economic and industrial optimization as outlined in the
previous section.
Managing disease risk
Disease remains a major loss factor in aquaculture, although Norwegian fish farming has successfully dealt with a number of health issues (Norwegian
Seafood Federation, 2010). Experience shows that new diseases emerge occasionally, and dealing with these is a big challenge.
It is still early days for integrated mathematical models of the immune system and its responses to pathogens, but the Virtual Physiological Human will
blaze the trail for this work too (e.g. Thomas et al., 2013). Consolidating in a Digital Salmon the growing understanding of immune responses is
key to being well prepared for dealing with novel diseases. Phenomics for monitoring immune responses to pathogen challenges are already in place, being
used to assess whether patients respond effectively to vaccines (Pulendran et al., 2010). Currently, systems vaccinology is mostly limited to
clustering analyses of multivariate immune responses, traced through gene regulation, protein expression and blood cell activity on time-scales from hours
to days. This high-dimensional phenotype already provides clues to physiological mechanisms by enabling comparison between individuals that respond well to
vaccination, non-responders, and healthy individuals. However, to follow up on these clues and realize the full potential of immune phenomics data requires
a physiome approach, seeking a systems understanding of how the immune system shapes its response to environmental perturbations by an interplay of system
state and immune processes.
Once the Digital Salmon can account for immune responses in terms of mechanism, it forms a framework for rapidly incorporating and interpreting new
knowledge such as massive data collection on diseased or vaccinated fish. Furthermore, practical experience with this approach has a good chance of
carrying over from one new disease to the next. Pioneering this systems approach with the Digital Salmon would also represent a substantial contribution to
general disease biology and Norway's visibility in this research arena.
Preserving wild salmon populations and the marine environment
By keeping the domesticated fish healthier on a more continuous basis, the knowledge base generated by a Digital Salmon Programme would represent a major
contribution to the preservation of wild salmon populations. It would also open for a deep systems understanding of the salmon’s neurophysiology that could
be used to modulate its neurobiological circuitry such that (possibly sterile) escapees do not move into the rivers. This would put an end to concerns
about genetic pollution as well as transmission of diseases in the rivers.
A systems understanding of neuroendocrine regulation and how the fish senses and responds to its environment (West-Eberhard, 2003; Gilbert & Epel,
2009; Bateson & Gluckman, 2011) will also greatly improve insight into the ecology of salmon, and thus its management outside the controlled conditions
of fish farms. In general, by making use of Digital Salmon models linking the genome and the phenome in a population dynamics and ecological context, we
would be in a much better position to understand how the genetic composition of wild salmon stocks would change as a function of a whole range of changes
in their freshwater and seawater habitats not necessarily imposed by the aquaculture industry, and how these changes would affect population numbers
(Hedger et al., 2013).
Thus, a Digital Salmon would become a very useful tool in ecosystem-based management of wild salmon populations, and it would tie into large-scale attempts
to model the general ecological dynamics in rivers and oceans. A Digital Salmon Programme should have these management aspects built into it.
Making a Norwegian Digital Salmon Programme a reality
The size of a Norwegian Digital Salmon Programme would have to be larger and differently organized than what can be accommodated through a few Centres of
Excellence or Centres for Research-based Innovation. But these concepts could very well be exploited under the umbrella of a programme structure that also
involves other mechanisms. A Norwegian Digital Salmon Programme would have to be run very differently from how large R&D programmes are currently run
by the Research Council of Norway. This does not imply that the current practice is not useful in some contexts, but a Norwegian Digital Salmon Programme
should ideally have the following characteristics:
-
Success criteria that are so explicitly stated that they can be used for programme assessment as well as provide clear guidance for defining
programme sub-goals throughout the whole programme period.
-
A flexible structure that stays in tune with international developments through very efficient procedures for monitoring programme-relevant
changes in technology, methodology and biological knowledge.
-
A coherent and closely monitored project portfolio where the outcomes of one project generation would at least in part provide the points of
departure for the next generation of projects.
-
An integrated and balanced mix of application-focused research and knowledge-oriented research, that not only fulfils overall practical goals but
also substantially improves Norway’s standing in the international research landscape.
-
Close links to international academic environments excelling in performing systems biology on model organisms and in systems medicine.
-
Openings for funding outstanding international groups to solve problems beyond the reach of Norwegian groups.
-
Opportunities for small start-up companies to develop crucial technology and methodology that could later become profitable products on
international biomedical markets.
-
A close interaction between universities, institute sector, industry, ministries and the Research Council of Norway promoting transparency,
division of labour and predictable funding premises allowing the R&D institutions to make long-term commitments.
-
A novel programme governance structure reflecting the need for concerted commitments of universities, institute sector, industry, ministries and
the Research Council of Norway.
Space restrictions prevent us from elaborating on all the above characteristics, and we therefore focus on points (iv), (vi), (vii), (viii) and (ix).
Integrating different types of research. It is well known that the considerable Norwegian investments in salmon aquaculture research have paid off remarkably poorly in major disciplinary arenas
in biology. In the fish biology sector instrumental research (application-focused work disregarding reasons behind phenomena) has been, and is still, the
favoured funding object for Norwegian authorities compared to more basic epistemic (knowledge-oriented) research. Instrumental research is likely to
continue enjoying the lion’s share of future funding unless specific measures are taken to balance the situation. Epistemic research is more highly valued
by the international research community and papers published in the high-impact journals in the main disciplinary categories, have in the majority of cases
a strong epistemic component. Considering the amount of money spent, Norway is not well represented in these journals. This is an unfortunate situation,
because it reflects suboptimal use of research money and negatively affects the quality of instrumental research activity, as it feeds heavily on knowledge
generated by curiosity-driven research. The Research Council of Norway is fortunately in a position to remedy this situation by demanding that researchers
running application-focused projects should articulate how their data also could be used to provide insights that would receive the attention of important
disciplinary journals.
The construction of an ensemble of mathematical descriptions of salmon physiology (including the genomic and environmental dimensions) by merging numerous
competences within the life sciences, the mathematical sciences and engineering, would take Norwegian interdisciplinary and interinstitutional
collaboration to an unprecedented level. The model construction as such would be a major knowledge-generator in itself. If we find that implementing
current conceptions of how a biological system or process works into a quantitative mathematical model that produce results in disagreement with existing
or new empirical data, this is a potent source for making new hypotheses about basic biological mechanisms which may result in important contributions to
the general biological research frontier. Moreover, the huge amounts of experimental data needed to both construct and test advanced models useful for
practical applications would be a treasure trove for discovering new basic patterns of phenomena in need of novel explanations. It would also drive the
development of new experimental protocols and new technology for measuring biological processes that would be of instrumental value for international
biological research in general. A Norwegian Digital Salmon Programme would thus blend application-focused research and knowledge-oriented research into a
concerted whole.
The US National Science Foundation defines transformative research as something that “involves ideas, discoveries, or tools that radically change
our understanding of an important existing scientific or engineering concept or educational practice or leads to the creation of a new paradigm or field of
science, engineering, or education. Such research challenges current understanding or provides pathways to new frontiers.” We anticipate that a Norwegian
Digital Salmon Programme would live up to this characteristic in several respects, and even become a role model for government-funded R&D by other
countries.
Funding international groups.
A Norwegian Digital Salmon Programme would need substantial help from international researchers due to reasons of intellectual and technological bandwidth.
Instead of just being based on current practice where Norwegian researchers invite international collaborators on their projects, we suggest that in order
to get the attention of the foremost world-wide competence, one should seriously consider establishing a mechanism for financing international
mission-critical R&D associated with the Digital Salmon. This could of course be a general mechanism for funding R&D in several fields of
particular interest to Norway. But it would be a particularly useful mechanism in connection with a vision-based and tightly governed biotechnology
programme like the Digital Salmon, where we want to achieve quite specific, measurable objectives by blending a wide array of competences within the life
sciences, the mathematical sciences and engineering.
The potential benefits would be great. We would substantially enhance the positive international attention for Norwegian research while tapping into
hundreds of billions NOK of invested capital in competence and infrastructure in other countries. Mission-critical goals would be achieved much faster
while not causing an imbalance between number of staff and students at the universities or overstaffing the institute sector. By demanding the
participation of Norwegian groups on highly specific terms in these projects, we would drive much larger portions of Norwegian research towards the
international elite level with regard to competence, professionalism, and participation in top-tier publications. This would substantially strengthen the
Norwegian group's competence, international visibility, and future attractiveness for international collaboration (e.g. with the EU or NIH/United States)
and recruitment of PhD students and postdocs. And we could demand a share for Norway in the profits from patentable inventions and other intellectual
property emerging from the projects financed.
Establishing new biotech industry. A Norwegian Digital Salmon Programme would demand development of a wide array of phenotyping technologies across the whole phenotypic hierarchy of
salmon, ranging from the intracellular level up to the whole individual. It would also demand innovative new software solutions for compiling and
analysing high-dimensional data and for linking such data to models. Finally, it would demand the evolution of advanced diagnostics tools, lab-on-a-chip
technologies, and new concepts for bioreactor design and synthetic biology engineering for producing salmon feed from new sustainable raw materials. As
these technologies would in many cases be directly applicable within biomedical R&D as well as in several other areas of the evolving bioeconomy, this
opens an opportunity window for funding start-up companies and established companies to solve mission-critical challenges that later could be introduced on
the international market. Such a mechanism would prevent breaking international rules while taking companies through the so-called “valley of death”,
where initial funding is running out while the product is not yet ready for the market.
Ensuring a concerted national effort.
As the Norwegian authorities have not so far defined a clear interface between the institute sector and the university sector in terms of funding
mechanisms and clear description of responsibilities (Kunnskapsdepartementet, 2009), it is quite daunting to understand how the authorities plan to realise
their dream about a much more pronounced collaboration, division of labour and concentration of efforts and resources (SAK) between and within these two
sectors. It is a frequent concern of the industry that much of the institute sector's R&D is not applied enough to really help the industry meet its
short-term goals, whereas the universities are lacking the basic competence needed to meet long-term goals characterized by very high innovation
thresholds. By having to merge, out of pure necessity, numerous competences within the life sciences, the mathematical sciences and engineering, a
Norwegian Digital Salmon Programme would in fact be an ideal instrument for realising, within a very important economic segment, a more pronounced SAK
structure. As the industry would also be on board and have a say in this structure, it would facilitate transparent commitments of the industry to
agreed-upon R&D goals to a much greater extent than is currently the case. But this would require more leadership by the authorities in connection with
programme design and role designation, and their willingness to smoothen the process by financial means.
In their book “Et kunnskapsbasert Norge” (A knowledge-based Norway), Torger Reve and Amir Sasson make a clear distinction between production of seafood and
biotechnology, which they intimately link to the diagnostics and pharmaceutical industries (Reve & Sasson, 2012). This distinction is not in resonance
with the fact that the Norwegian salmon production industry is indeed already a biotechnological industry according to the OECD definition. If Reve and
Sasson had acknowledged this before discussing the prospects for developing a biotech industry in Norway, it would probably have affected their conclusion.
A Norwegian Digital Salmon Programme would likely have a very positive impact with regard to several of the assessment criteria Reve and Sasson use to
classify and qualify the Norwegian knowledge industries: cluster attractivity, educational attractivity, talent attractivity, research and innovation
attractivity, ownership attractivity, and environmental attractivity. These aspects should be given close attention when designing the Digital Salmon
Programme. A successful design could make this programme a driver for the creation of a biotechnological supplier industry that in addition to serving the
Norwegian salmon production companies, would find large international markets in a wide array of segments also outside aquaculture for their products. This
would echo what has happened in the Norwegian petroleum sector, where the supplier industry now delivers advanced technology, products and services for the
Norwegian shelf as well as international markets. This success story is very much due to close interaction between the oil companies on the shelf, the
supplier industry and the R&D environments.
From the 1920s through the 1980s, Bell Labs, the R&D wing of AT&T, was the most innovative scientific organization in the world. As Jon Gertner
argues in his new book, “The Idea Factory”, it was where the future was invented (Gertner, 2012). Bell Labs was behind many of the innovations that have
come to define modern life, including the transistor, the laser, the silicon solar cell, the communication satellite, the cellular telephone system and the
fibre-optic cable system. Besides access to talent, this success story was based on visionary leadership, forced interdisciplinarity, purpose-guided
freedom and institutional patience. Nothing prevents Norway from trying out this recipe to create transformative technology through a Digital Salmon
Programme as envisioned above.
In his delightful book “The Black Swan”, Nassim N. Taleb distinguishes between two realms, Mediocristan and Extremistan (Taleb, 2007). Mediocristan is
where normality resides and the impacts of events are rather small. In Extremistan, events that seemed unlikely, impossible and even unthinkable occur
frequently and have a dramatic impact. Black Swan events occur in Extremistan. Taleb’s point is that we should pay much higher attention to the possibility
of Black Swan events when we assess the future prospects of things. One can very well make a linear projection of future production volumes of salmon in
2050 from current volumes, but this is a daunting thing to justify, considering the harshness of the Darwinian theatre that the salmon production industry
is part of. We would be unbelievably lucky if no Black Swan with dramatic negative impact appeared within the next 40 years if we continue with business as
usual (see also Chapter 5 by Tveterås in this book). There are many situations we cannot prepare for, but when it comes to biology, a Digital Salmon
Programme would prepare us for a whole range of events that otherwise would develop into Black Swan events.
There is no excuse for not starting preparing for the highly unexpected in terms of building a real pre-emptive knowledge base for salmon biology.
Considering that Norway has invested several billions NOK in CO2-handling R&D, there is apparent willingness to take on risky projects when
this is found opportune by those controlling the expenditure of Norwegian oil money. Even without any expansion of volumes, we will from now till 2050
produce 40 million tons with a current market value of >1200 billion NOK. The strong possibility of not being able to realise this value due to lack of
knowledge, should be enough reason for spending several billion NOK on a Norwegian Digital Salmon Programme that at the same time would give broad segments
of Norwegian science a real boost.
Acknowledgements
We are grateful to Petter Arnesen, Kjetil Hindar, Ole Petter Ottersen, Karl Shearer, Trygve Sigholt, Per Olav Skjervold and Dag Inge Våge for their
perceptive comments on the issues discussed here. Figure 3 is based on two Creative Commons licensed images (Kils, 2005; Rajasingh et al., 2006,
Figure 1); the composite image is available at http://arken.nmbu.no/~jonvi/digital_salmon.jpg.
References
Anon (2010). QTL Eggs – Documented IPN protection from day one. Aqua Gen Information Letter 2010, 1.
Barrows FT, Bellis D, Krogdahl Å, Silverstein JT, Herman EM, Sealey WM, Rust MB & Gatlin DM (2008). Report of the Plant Products in Aquafeed Strategic
Planning Workshop: An integrated, interdisciplinary research roadmap for increasing utilization of plant feedstuffs in diets for carnivorous fish. Reviews in Fisheries Science 16, 449–455.
Bateson P & Gluckman P (2011). Plasticity, Robustness, Development and Evolution. Cambridge University Press.
Beard DA, Neal ML, Tabesh-Saleki N, Thompson CT, Bassingtwaighte JB, Shimoyama M & Carlson BE (2012). Multiscale Modeling and Data Integration in the
Virtual Physiological Rat Project. Annals of Biomedical Engineering 40, 2365–2378.
De Bono B, Hoehndorf R, Wimalaratne S, Gkoutos G & Grenon P (2011). The RICORDO approach to semantic interoperability for biomedical data and models:
strategy, standards and solutions. BMC Research Notes 4, 313.
Carroll L (1871). Through the Looking-Glass. Project Gutenberg, http://www.gutenberg.org/ebooks/12.
FAO (2006). Livestock’s Long Shadow: Environmental Issues and Options. Food and Agriculture Organization of the United Nations.
Gertner J (2012). The idea factory: Bell Labs and the great age of American innovation. Penguin Press.
Gilbert SF & Epel D (2009). Ecological Developmental Biology. Sinauer Associates.
Gjuvsland AB, Vik JO, Beard DA, Hunter PJ & Omholt SW (2013). Bridging the genotype-phenotype gap: what does it take? The Journal of Physiology; DOI: 10.1113/jphysiol.2012.248864.
Gove PB ed. (1981). Webster’s Third New International Dictionary of the English Language, Unabridged. Merriam, Springfield, MA.
Gurwitz D (2012). High-Quality Phenomics are Crucial for Informative Omics Studies. Drug Development Research 73, 353–356.
Hedger RD, Sundt-Hansen LE, Forseth T, Ugedal O, Diserud OH, Kvambekk ÅS, Finstad AG & Bradford M (2013). Predicting climate change effects on
subarctic–Arctic populations of Atlantic salmon (Salmo salar). Canadian Journal of Fisheries and Aquatic Sciences 70, 159–168.
Houle D, Govindaraju DR & Omholt S (2010). Phenomics: the next challenge. Nat Rev Genet 11, 855–866.
Hunter P et al. (2013). A vision and strategy for the virtual physiological human: 2012 update. Interface Focus; DOI:
10.1098/rsfs.2013.0004.
Hunter PJ & Borg TK (2003). Integration from proteins to organs: the Physiome Project. Nat Rev Mol Cell Biol 4, 237–243.
Joyce AR & Palsson BØ (2006). The model organism as a system: integrating “omics” data sets. Nature Reviews Molecular Cell Biology 7, 198–210.
Kils U (2005). File:Salmonlarvakils.jpg. Wikipedia. Available at:
http://en.wikipedia.org/w/index.php?title=File:Salmonlarvakils.jpg&oldid=482663815 [Accessed April 29, 2013].
Krogdahl Å, Penn M, Thorsen J, Refstie S & Bakke AM (2010). Important antinutrients in plant feedstuffs for aquaculture: an update on recent findings
regarding responses in salmonids. Aquaculture Research 41, 333–344.
Kunnskapsdepartementet (2009). Klima for forskning. St.meld. nr. 30 (2008-2009). Available at:
http://www.regjeringen.no/nb/dep/kd/dok/regpubl/stmeld/2008-2009/stmeld-nr-30-2008-2009-.html?id=556563 [Accessed March 3, 2013].
Land S, Niederer SA, Louch WE, Sejersted OM & Smith NP (2013). Integrating multi-scale data to create a virtual physiological mouse heart. Interface Focus; DOI: 10.1098/rsfs.2012.0076.
Lanktree MB, Hassell RG, Lahiry P & Hegele RA (2010). Phenomics: expanding the role of clinical evaluation in genomic studies. Journal of Investigative Medicine 58, 700–706.
Lieschke GJ & Currie PD (2007). Animal models of human disease: zebrafish swim into view. Nat Rev Genet 8, 353–367.
Merriam-Webster (2008). Webster’s Third New International Dictionary, Unabridged. Available at: http://unabridged.merriam-webster.com.
Morais S, Silva T, Cordeiro O, Rodrigues P, Guy DR, Bron JE, Taggart JB, Bell JG & Tocher DR (2012). Effects of genotype and dietary fish oil
replacement with vegetable oil on the intestinal transcriptome and proteome of Atlantic salmon (Salmo salar). BMC Genomics 13,
448.
Naylor RL, Hardy RW, Bureau DP, Chiu A, Elliott M, Farrell AP, Forster I, Gatlin DM, Goldburg RJ, Hua K & Nichols PD (2009). Feeding aquaculture in an
era of finite resources. PNAS 106, 15103–15110.
Niederer SA, Fink M, Noble D & Smith NP (2009). A meta-analysis of cardiac electrophysiology computational models. Experimental Physiology 94, 486–495.
Norwegian Seafood Federation (2010). Health situation of farmed salmonids since 1990. Norwegian Seafood Federation Factsheets. Available at:
http://www.fhl.no/getfile.php/DOKUMENTER/Faktaark_laksesykdommer%201990%20-2009ENG.pdf [Accessed March 6, 2013].
Noy NF, Shah NH, Whetzel PL, Dai B, Dorf M, Griffith N, Jonquet C, Rubin DL, Storey M-A, Chute CG & Musen MA (2009). BioPortal: ontologies and
integrated data resources at the click of a mouse. Nucl Acids Res 37, W170–W173.
OECD (2005). Statistical Definition of Biotechnology. Available at: http://www.oecd.org/sti/biotech/statisticaldefinitionofbiotechnology.htm [Accessed
April 26, 2013].
Olafsen T, Winther U, Olsen Y & Skjermo J (2012). Value created from productive oceans in 2050. Report of the Royal Norwegian Society of Sciences and Letters and of the Norwegian Academy of Technological Sciences. Available at:
http://www.sintef.no/home/Fisheries-and-Aquaculture/News/Value-created-from-productive-oceans-in-2050/ [Accessed March 3, 2013].
Olsen OA (1989). Structured modelling of fish physiology. Dr. Ing. dissertation 1989:37, Norwegian Technical University, Department of technical
cybernetics, Trondheim.
Omholt SW (2012). From sequence to consequence and back. Progress in Biophysics and Molecular Biology; DOI: 10.1016/j.pbiomolbio.2012.09.003.
Pulendran B, Li S & Nakaya HI (2010). Systems Vaccinology. Immunity 33, 516–529.
Rajasingh H, Gjuvsland AB, Våge DI & Omholt SW (2008). When parameters in dynamic models become phenotypes: a case study on flesh pigmentation in the
chinook salmon (Oncorhynchus tshawytscha). Genetics 179, 1113–1118.
Rajasingh H, Øyehaug L, Våge DI & Omholt SW (2006). Carotenoid dynamics in Atlantic salmon. BMC Biology 4, 10.
Reve T & Sasson A (2012). Et kunnskapsbasert Norge. Universitetsforlaget, Oslo, Norway. Available at:
http://www.bokkilden.no/SamboWeb/produkt.do?produktId=5774939 [Accessed April 3, 2013].
Taleb NN (2007). The Black Swan: The Impact of the Highly Improbable. Penguin Press.
Thomas N, Heather J, Pollara G, Simpson N, Matjeka T, Shawe-Taylor J, Noursadeghi M & Chain B (2013). The immune system as a biomonitor: explorations
in innate and adaptive immunity. Interface Focus; DOI: 10.1098/rsfs.2012.0099.
Torrissen O, Olsen RE, Toresen R, Hemre GI, Tacon AGJ, Asche F, Hardy RW & Lall S (2011). Atlantic salmon (Salmo salar): The “super-chicken” of the
sea? Reviews in Fisheries Science 19, 257–278.
Vik JO, Gjuvsland AB, Li L, Tøndel K, Niederer SA, Smith NP, Hunter PJ & Omholt SW (2011). Genotype–phenotype map characteristics of an in silico heart
cell. Frontiers in Physiology 2, 106.
Wacyk J, Powell M, Rodnick K, Overturf K, Hill RA & Hardy R (2012). Dietary protein source significantly alters growth performance, plasma variables
and hepatic gene expression in rainbow trout (Oncorhynchus mykiss) fed amino acid balanced diets. Aquaculture 356–357, 223–234.
Weinstein JA, Jiang N, White RA, Fisher DS & Quake SR (2009). High-Throughput Sequencing of the Zebrafish Antibody Repertoire. Science 324, 807–810.
West-Eberhard MJ (2003). Developmental Plasticity and Evolution. Oxford University Press, USA.
Wiktionary (2013). Wiktionary. Available at: http://en.wiktionary.org [Accessed June 20, 2013].
Øverland M, Tauson A-H, Shearer K & Skrede A (2010). Evaluation of methane-utilising bacteria products as feed ingredients for monogastric animals. Archives of Animal Nutrition 64, 171–189.




