Master projects on genome-scale metabolic modeling at NMBU
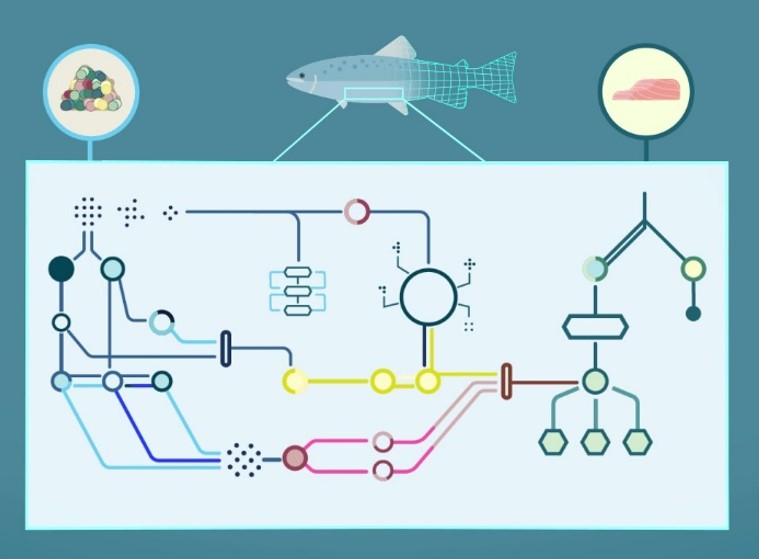
How does a salmon and the microbes in its gut convert nutrients from the feed into the fillet that we eat?
To answer such questions, we use mathematical models to study the metabolic networks of enzymatic reactions that are encoded in genomes.
Master projects are available within three research areas: feed prediction for salmon, integration of models and omics data, and development of scalable modeling tools. You are also highly encouraged to come to us with your own ideas!
All projects aim to answer interesting biological questions by combining models and data and require basic knowledge of biochemistry, programming, and linear algebra. We mainly use Python and the toolbox COBRApy, which makes it easy to get started with modeling.
The main supervisor is Ove Øyås who is a postdoc working with Jon Olav Vik and Filip Rotnes in the Bioinformatics and Applied Statistics (BIAS) group. We collaborate with other experimental and modeling groups at NMBU and internationally.
Master projects with Jon Olav as main supervisor are available here.
Research areas
Predicting new feeds for salmon
Atlantic salmon (Salmo salar) is the world's most valuable fish commodity and Norway is the largest producer. Increased demand for feed and insufficient marine resources has led to a switch to plant ingredients, but current plant-based feeds have a negative impact on fish health and the environment. Therefore, finding feeds that minimize cost and environmental impact while providing necessary nutrients to the fish is one of the key challenges facing the industry. In your master project, you can help us address this challenge by using metabolic models of salmon and its gut microbes to predict new plant-based feeds based on renewable resources. These projects are suitable for students from all backgrounds.
Integrating models and omics data
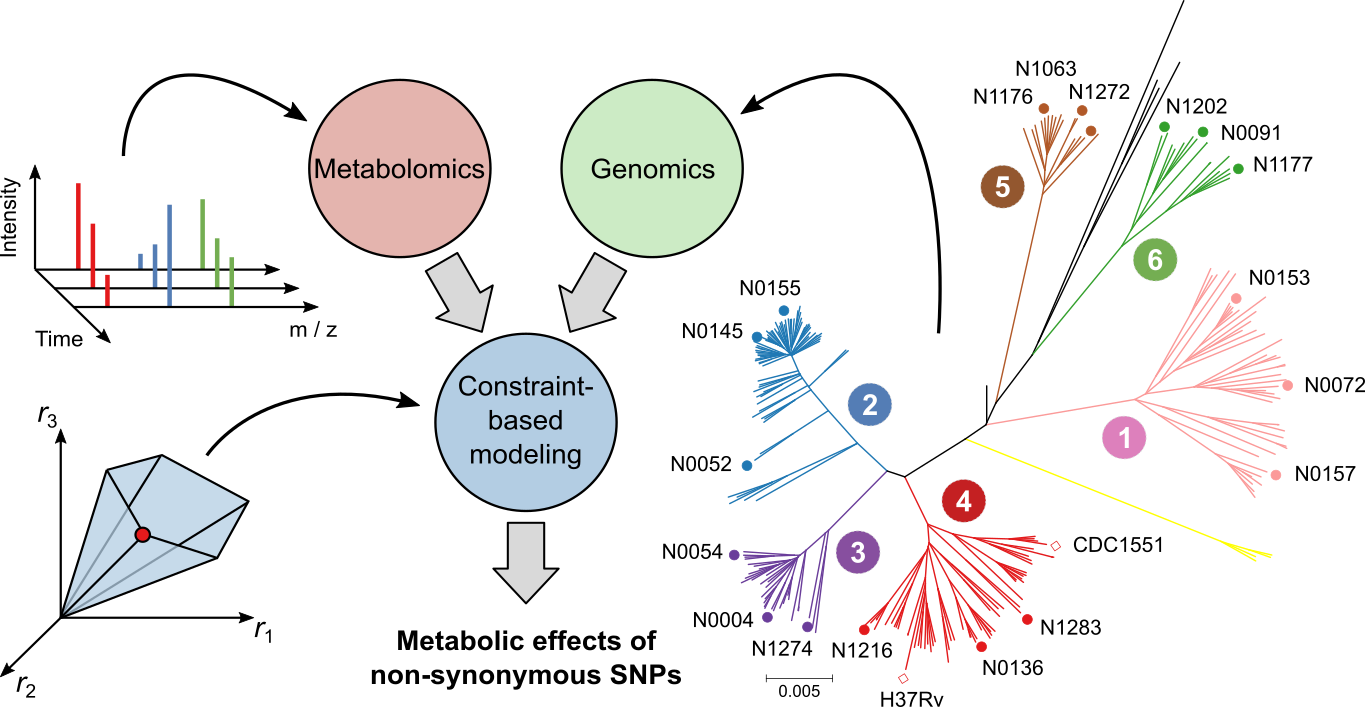
High-throughput omics technologies are advancing rapidly, producing large data sets that aim to capture the entire genome, transcriptome, proteome, or metabolome of an organism. However, integration with mathematical models is needed in order to make sense of the accumulating data. We are using metabolic modeling to integrate omics data from salmon and its gut microbiota, aiming to identify and explain observed differences between fish, tissues, and experimental conditions. You can help us transform omics data into valuable insights for salmon biology and aquaculture by applying state-of-the-art computational methods. These projects are suitable for students who have some experience with programming and data analysis.
Developing scalable modeling tools
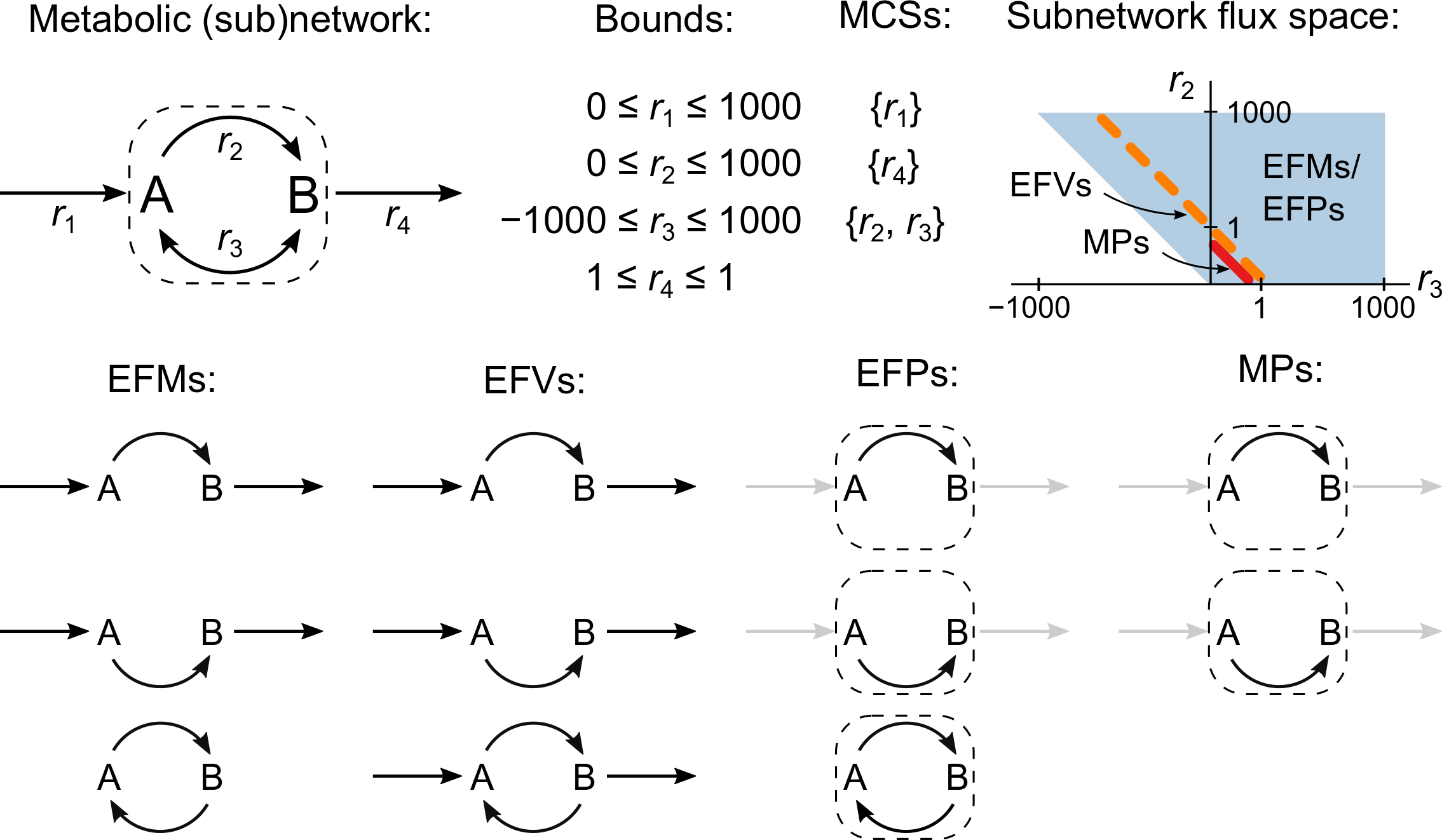
Metabolic models are available for a wide range of organisms and their scope of application is expanding from individual cells to multicellular systems such as microbial communities or animal tissues. However, the computational cost of many key analysis methods grows exponentially with network size, increasing the need for new modeling tools that scale to large networks. You can help us develop scalable tools that address this fundamental challenge and make it possible to answer new biological questions. These projects are suitable for students with good knowledge of programming and mathematics and an interest in biology (e.g. engineers).
Contact
Don't hesitate to contact us if you are interested in learning more about the projects! Send an email or drop by office M251.